Colossal scientists used CRISPR technology, a gene-editing tool, to modify the genetic material of the gray wolf and replace them with dire wolf traits found in the ancient DNA.
Gene editing for de-extinction is a cutting-edge area of biotechnology that aims to bring extinct species back to life—or at least re-create close genetic approximations of them—using tools like CRISPR-Cas9 and cloning.
cutting-edge genetic engineering strategies employed by Colossal Biosciences in their mission to de-extinct the dire wolf (Aenocyon dirus), a Pleistocene apex predator. Utilizing CRISPR-Cas9, a revolutionary gene-editing system, researchers aim to integrate functional genomic sequences from ancient dire wolf DNA into the genome of the modern gray wolf (Canis lupus). This work not only advances the frontier of synthetic biology and conservation genetics but also raises novel questions about the feasibility and implications of ecological restoration via genetic proxy.
How Gene Editing Enables De-Extinction
1. DNA Recovery
Scientists first recover DNA from preserved remains of extinct species (like frozen mammoths or museum specimens). The older the DNA, the more degraded it tends to be, so this step is difficult and sometimes only partial genomes can be reconstructed.
2. Genome Sequencing and Comparison
The extinct species’ genome is compared to that of a close living relative. For instance, the woolly mammoth is compared to the Asian elephant, and the passenger pigeon to the band-tailed pigeon.
3. CRISPR Gene Editing
CRISPR allows scientists to “cut and paste” genes from the extinct species into the genome of the living relative. This can create a genetically modified organism that exhibits traits of the extinct species (e.g., thick fur, fat storage, cold resistance in mammoths).
4. Cell Culturing and Cloning
The edited DNA is inserted into an egg cell (often from the modern relative), which is then implanted in a surrogate mother, or grown using advanced reproductive techniques (like stem cell differentiation).
Projects:
Colossal Biosciences: Focused on reviving the woolly mammoth and the thylacine (Tasmanian tiger). Their goal is not just to bring these animals back, but to reintroduce them to their native ecosystems to help restore ecological balance.
he dire wolf, once a dominant predator across North America, went extinct approximately 10,000 years ago. Long thought to be a close relative of the modern gray wolf, recent paleogenomic analyses revealed that dire wolves diverged from the lineage of Canis over 5 million years ago, positioning them in a distinct evolutionary genus: Aenocyon.
Despite this divergence, the modern gray wolf is the closest living physiological analog, making it the ideal biological chassis for attempting de-extinction through genome engineering. Colossal Biosciences has taken on the challenge, employing CRISPR-Cas9 and advanced comparative genomics to reintroduce key dire wolf traits into the gray wolf genome.
Ancient DNA Extraction and Sequencing
Colossal’s work began with the acquisition of ancient DNA (aDNA) samples from well-preserved dire wolf fossils found in La Brea Tar Pits, California. Due to postmortem degradation and cytosine deamination, specialized clean room protocols and next-generation sequencing (NGS) techniques were used to reconstruct a partial but high-confidence dire wolf genome.
Bioinformaticians employed genome alignment algorithms to map recovered dire wolf sequences against the high-quality Canis lupus reference genome (CanFam4). From this alignment, researchers identified approximately 18,000 SNPs, 450 indels, and several large structural variants representing candidate regions for phenotypic divergence.
Revive & Restore: A nonprofit leading efforts to bring back the passenger pigeon and the heath hen, among others, with an emphasis on conservation outcomes.
Trait Selection for Gene Editing
Based on fossil and isotopic data, Colossal focused on a suite of phenotypic traits that defined dire wolves:
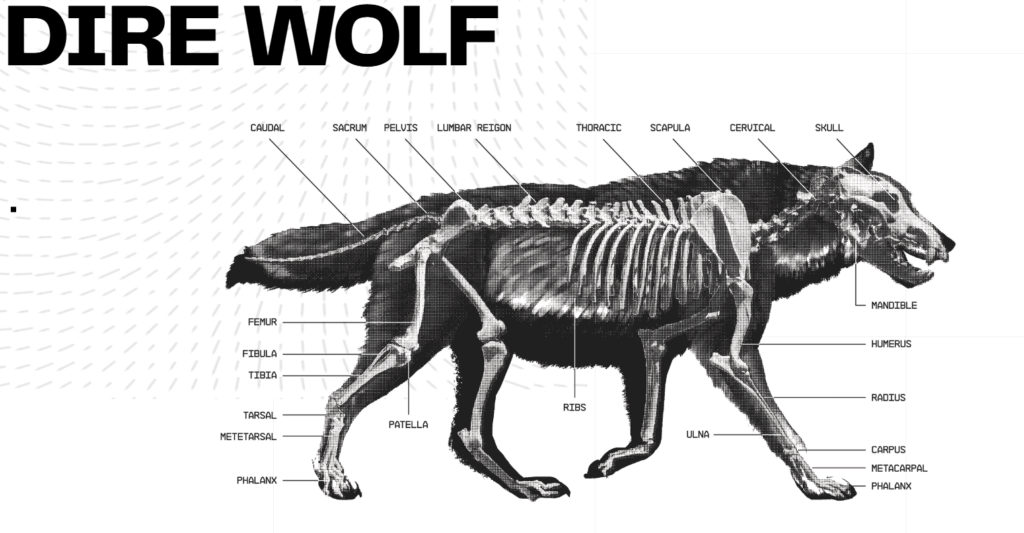
- Cranial Robusticity: Large, wide skulls capable of high bite force.
- Hypercarnivorous Dentition: Enhanced molar shearing surfaces.
- Cold Adaptation: Variants in UCP1 and mitochondrial genes.
- Large Body Size: Growth regulation genes including IGF1, GHR, and HMGA2.
Using comparative genomics and evolutionary constraint analyses (e.g., PhyloP, GERP scores), they filtered for high-impact coding variants and regulatory elements likely responsible for these traits.
CRISPR Design and Integration Strategy
Using a multiplexed CRISPR-Cas9 approach, Colossal scientists designed guide RNAs (gRNAs) to target over 60 genomic loci in the gray wolf genome for replacement or editing.
Editing Strategy:
- HDR (Homology-Directed Repair) was used to swap in dire wolf alleles using synthetic donor templates.
- Base editing and prime editing were employed for single-nucleotide changes, especially in non-coding regulatory elements.
- Lentiviral and electroporation delivery systems were optimized in vitro using induced pluripotent stem cells (iPSCs) derived from gray wolf fibroblasts.
Custom Cas9 variants with reduced off-target activity (e.g., SpCas9-HF1) were used to ensure high precision, and computational prediction tools like CRISPOR and CCTop were employed to minimize unwanted edits.
Synthetic Embryogenesis and Embryo Transfer
The edited gray wolf iPSCs were differentiated into gametes using organoid culture techniques, and fertilized in vitro to produce synthetic embryos. These embryos were implanted into surrogate gray wolf females under strict veterinary supervision.
Embryogenesis monitoring used CRISPR lineage tracing and single-cell RNA sequencing to ensure the development of dire wolf-like morphogenesis, particularly in limb, craniofacial, and neural crest derivatives.
Validation and Characterization
Following live births, Colossal employed a multi-tiered phenotyping strategy:
- Genomics: Whole-genome sequencing confirmed intended edits and ruled out off-target mutations.
- Transcriptomics: RNA-Seq of blood, skin, and neural tissues verified expression profiles consistent with reconstructed dire wolf regulatory networks.
- Morphometrics: CT scans and morphometric analysis of skulls and limbs demonstrated enhanced cranial width and limb proportions matching dire wolf fossil data.
- Behavioral assays: Ethologists assessed prey response, social dynamics, and cold climate behavior in semi-wild enclosures.
Ecological and Ethical Implications
While the resulting animals are not true dire wolves in the strict taxonomic sense, they represent a functional proxy—a genomic and phenotypic composite capable of occupying the ecological niche of the extinct dire wolf.
However, such interventions raise major ethical concerns:
- Genetic integrity of existing species
- Animal welfare of engineered individuals
- Ecological unpredictability of reintroductions
Colossal collaborates with conservation ecologists to model ecosystem impacts before any potential field release.
By leveraging CRISPR technology, Colossal has demonstrated the feasibility of trait-level de-extinction, blurring the lines between synthetic biology and conservation. While challenges remain, this project establishes a blueprint for de-extincting highly divergent species through modular genome engineering and targeted phenotypic reconstruction.
