CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology has revolutionized genetic editing across various organisms, including plants. Here’s how CRISPR is utilized in plant genetic editing:
- Targeted Gene Editing: CRISPR enables precise modifications in the plant genome by targeting specific DNA sequences. Researchers design guide RNA molecules complementary to the target DNA sequence, which then guides the CRISPR-associated protein (Cas) to the desired location in the genome, where it can induce changes like insertions, deletions, or replacements in the DNA sequence.
- Trait Modification: CRISPR technology can be employed to introduce or modify specific traits in plants. This includes enhancing disease resistance, improving tolerance to environmental stresses such as drought or extreme temperatures, increasing yield, altering nutritional content, or modifying other agronomically desirable traits.
- Accelerated Breeding: Traditional breeding methods can take years to produce desired traits in plants. CRISPR allows for faster and more precise modifications, significantly accelerating the breeding process. This can lead to the development of new plant varieties with improved characteristics in a shorter time frame.
- Reduced Off-Target Effects: While no genetic editing method is completely error-free, CRISPR has been refined to minimize off-target effects, where unintended changes occur in the genome. This precision is crucial for ensuring the safety and stability of genetically edited plants.
- Functional Genomics: CRISPR technology facilitates the study of gene function in plants by enabling targeted gene knockouts or knock-ins. By disrupting or introducing specific genes, researchers can elucidate the roles of these genes in various biological processes, providing valuable insights into plant biology and potential targets for crop improvement.
- Regulatory Considerations: The regulatory landscape surrounding CRISPR-edited plants varies by country. Some jurisdictions have classified certain CRISPR-edited plants as non-GMOs (genetically modified organisms) if they do not contain foreign DNA. This distinction has implications for the regulation and commercialization of CRISPR-edited crops.
CRISPR revolution began when the research teams from the US and Europe led by Jennifer Doudna and Emmanuelle Charpentier revealed how this bacterial defense system could be turned into a phenomenal genome-editing tool. The two scientists studied the CRISPR system in Streptococcus pyogenes, a gram-positive pathogenic bacterium with Cas9 protein involved in its adaptive immune system.
The Cas9 (CRISPR associated protein 9) is a DNA endonuclease enzyme associated with CRISPR adaptive immunity in S. pyogenes. Cas9 is an RNA-guided endonuclease and thus can cleave nearly any sequence of DNA complementary to the guide RNA.
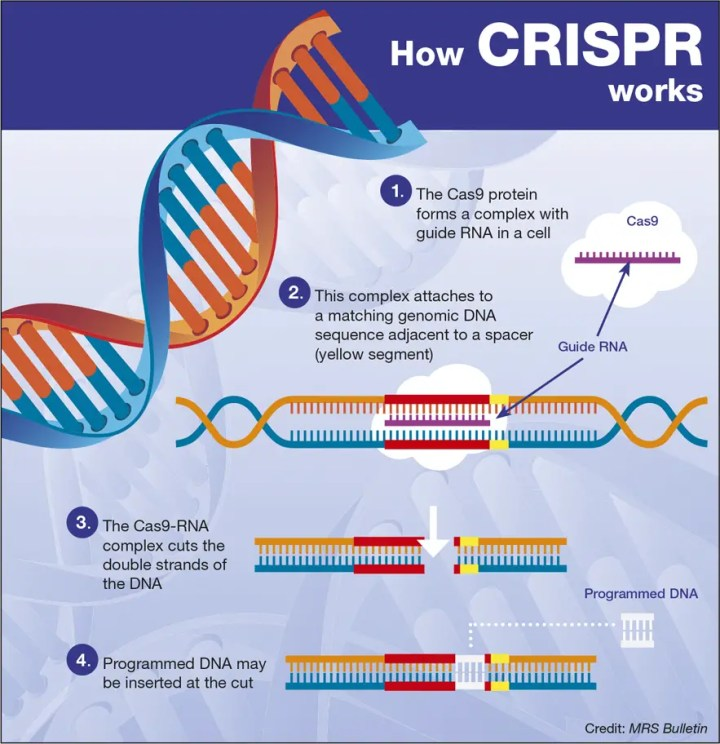
Till now conventional breeding techniques have been extensively used to improve the yield and agronomic performance which is a complex, time-consuming and labor-intensive process (Zhang et al., 2018). It is a selection of improved individuals utilizing genetic variation in the population (Breseghello and Coelho, 2013). It also recombines the desired gene pools resulting in new genotypes or cultivars (Holme et al., 2019). However, when the desirable genetic variation is not available in the gene pool, then it can be generated and used for selection by inducing mutations using various mutagenic agents like non-ionizing radiation i.e. UV rays or ionizing radiation i.e. X and gamma rays, alpha and beta rays, fast and slow neutrons. Some chemicals can also be used as mutagenic agents like ethyl methane sulphonate (EMS), methyl methane sulphonate (MMS), hydrogen fluoride (HF), sodium azide, N-methyl-N-nitrosourea (MNU) and hydroxylamine) (Parry et al., 2009). Mutation breeding has not been extensively utilized in vegetable crop improvement except for a few exceptions being low in its efficiency. Over the last few decades, there have been a large number of significant developments in the molecular biology approaches to improve crop yield and quality. Recently, tremendous progress has been made in genome editing tools like site-directed nucleases (SDNs) which are able to edit the crops at high speed and possess great potential in shaping up the novel genetic makeup of vegetable crops (Tian et al., 2021). Genome editing tools can precisely engineer the genes by either deleting, replacing or inserting specific sequences at the specific targeted location in the target genome to generate novel traits (Corte et al., 2019). Zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated enzymes are the tools for genome editing used to modify plants (Miller et al., 2007). Site-specific double-stranded breaks (DSBs) are enabled by Crispr/Cas which further activate the cellular DNA repair systems (Zhu et al., 2020; Gaj et al., 2013). These DSBs can either be corrected by the non-homologous end joining (NHEJ) pathway or through the homology-directed repair (HDR) pathway (O’Driscoll and Jeggo, 2006; Wang T. et al., 2019a). The use of first-generation technologies like ZFNs and TALENs has been limited due to their adverse mutagenic outcome, low editing efficiency, time-consuming process and labor-intensive selection and screening process (Gaj et al., 2013; Jaganathan et al., 2018). The second-generation genome editing technology i. e. CRISPR/Cas9 is easier to design, and execute and more cost-effective. The use of CRISPR/Cas9 in vegetable crops has substantially expanded gene editing technology and made it possible to create novel genotypes with desired phenotypic features and altered genomic functions at the base pair level (Abdallah et al., 2015; Nunez de Caceres Gonzalez and De la Mora Franco, 2020). We will first go over CRISPR/Cas9 history and development before summarising how it is currently used to modify vegetable crops. Finally, we will talk about the real-world challenges in enhancing vegetable crops with the desired traits.
Clustered regularly interspaced short palindromic repeats-CRISPR-associated protein-9
CRISPR-Cas9 is an advanced genome editing technique that enables scientists to change, add, or remove specific DNA sequences to modify specific regions of the genome. In general, there are three main types (I-III) of CRISPR-Cas systems utilized for target interference (Rouillon et al., 2013). Type II uses its two distinctive nuclease domains, RuvC and HNH, to achieve interference with only a basic effector-module design (Gasiunas et al., 2012). Type II Cas9 from Streptococcus pyogenes (SpCas9) is the most popular CRISPR nuclease employed in CRISPR-Cas technology (Doudna and Charpentier, 2014). The protospacer adjacent motif (PAM) is recognized by the sgRNA-Cas complex, and Cas9 cleaves the target DNA to create a double-strand break (DSB), activating cellular DNA repair processes.
